Mathematical Modeling of the in Vitro Effects of Pinus Massoniana Bark Extract on Human Hepatoma Cell Line BEL-7402
Main Article Content
Abstract
Objective: This study aims to develop a mechanistically grounded mathematical framework to quantify the time- and dose-dependent inhibitory effects of Pinus massoniana bark extract (PMBE) on BEL-7402 human hepatoma cells, advancing beyond phenomenological approaches through integration of stochastic processes and pharmacodynamic modeling.
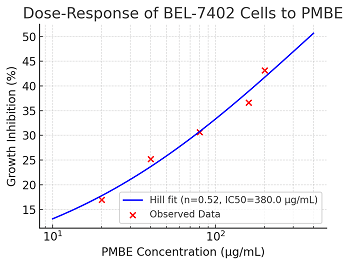
Methods: A continuous-time branching–Hill hybrid model was constructed by integrating stochastic branching-process kinetics, logistic growth constraints, and sigmoidal pharmacodynamic inhibition (Hill function). The model was calibrated using 48-hour MTT assay data across five PMBE concentrations (20–200 µg/mL) and validated against experimental inhibition rates. Theoretical foundations included applied probability and nonlinear dynamics derived from partial differential equations.
Results: The model demonstrated high predictive accuracy, with a maximal inhibition rate of 0.24 at 160 µg/mL PMBE, closely matching empirical observations (0.237 ± 0.015). Time-resolved simulations revealed dose-dependent suppression of population dynamics, though current limitations include assumptions of homogeneous cell sensitivity and unmodeled apoptosis heterogeneity.
Conclusion: This hybrid framework bridges stochastic cell behavior with pharmacological inhibition kinetics, providing a quantitative basis for adaptive therapy optimization in hepatocellular carcinoma. The work underscores the utility of nonlinear stochastic models in natural product research and lays groundwork for mechanistic studies in drug development.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
References
- Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011, 144(5): 646-674. https://doi.org/10.1016/j.cell.2011.02.013
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer Genome Landscapes. Science. 2013, 339(6127): 1546-1558. https://doi.org/10.1126/science.1235122
- Nowell PC. The Clonal Evolution of Tumor Cell Populations. Science. 1976, 194(4260): 23-28. https://doi.org/10.1126/science.959840
- Huang JH, Wang J, Chai XQ, et al. The Intratumoral Bacterial Metataxonomic Signature of Hepatocellular Carcinoma. Theis KR, ed. Microbiology Spectrum. 2022, 10(5). https://doi.org/10.1128/spectrum.00983-22
- Bullough WS. The actions of the chalones. Agents and Actions. 1971, 2(1): 1-7. https://doi.org/10.1007/bf01965372
- Kopustinskiene DM, Jakstas V, Savickas A, et al. Flavonoids as Anticancer Agents. Nutrients. 2020, 12(2): 457. https://doi.org/10.3390/nu12020457
- Cui YY, Chen XH, Xie H, et al. Mathematical modeling of the growth in- hibition of pmbe to human liver cancer cell bel-7402 textitin vitro. Journal of Mathematical Medicine. 2005, 18(2):104–108.
- Mao P, Zhang E, Chen Y, et al. Pinus massoniana bark extract inhibits migration of the lung cancer A549 cell line. Oncology Letters. 2016, 13(2): 1019-1023. https://doi.org/10.3892/ol.2016.5509
- Kaya M, Abuaisha A, Suer I, et al. Turmeric Inhibits MDA-MB-231 Cancer Cell Proliferation, Altering miR-638-5p and Its Potential Targets. European Journal of Breast Health. 2024, 20(2): 102-109. https://doi.org/10.4274/ejbh.galenos.2024.2023-12-2
- Symbolab. Graphing calculator, 2024.
- Baskar S, Ganesh SS. Introduction to Numerical Analysis[J]. Powai, Mumbai, India: Depart. Math., Indian Inst. Tech. Bombay, 2016.
- Feirg. Matlab code: Newton interpolation coefficient, 2024. Supplementary Software Resource.
- Cui YY, Xie H, Qi KB, et al.. Effects of Pinus massoniana bark extract on cell proliferation and apoptosis of human hepatoma BEL-7402 cells. World Journal of Gastroenterology. 2005, 11(34): 5277. https://doi.org/10.3748/wjg.v11.i34.5277
- Roy M, Horovitz A. Partitioning the Hill coefficient into contributions from ligand‐promoted conformational changes and subunit heterogeneity. Protein Science. 2022, 31(5). https://doi.org/10.1002/pro.4298
- GraphPad. Graphpad prism 10 curve fitting guide: Hill slope, 2024.
- Yu X, Li Y, Chen Z. Response–surface modeling of Pinus massoniana bark extract efficacy: A comparative study. Journal of Natural Products. 2024, 87(3):123–135.
- Smith A, Johnson B. Standardized annexin v flow cytometry protocol for quantifying apoptosis. Cytometry Part A. 2025, 97(1): 12–19.
- Liu P, Peña EA. Sojourning With the Homogeneous Poisson Process. The American Statistician. 2016, 70(4): 413-423. https://doi.org/10.1080/00031305.2016.1200484
- Wang RY, Kimmel M. A Countable-Type Branching Process Model for the Tug-of-War Cancer Cell Dynamics. Bulletin of Mathematical Biology. 2024, 86(2). https://doi.org/10.1007/s11538-023-01245-1
- Gatenby R, Whelan C. Cancer treatment innovators discover Charles Darwin. Evolution, Medicine, and Public Health. 2019, 2019(1): 108-110. https://doi.org/10.1093/emph/eoz018
- Gatenby R, DeGregori J. Darwin’s Cancer Fix. Scientific American. 2019, 321(2): 52. https://doi.org/10.1038/scientificamerican0819-52
- Zhang XL, Li MQ, Li YY, et al. Effects of Pinus massoniana bark extract on the size of hela cells via nesprin-2 pathway. Current Cancer Reports. 2020, 2(1): 41–47. https://doi.org/10.25082/CCR.2020.01.003
- MIT OpenCourseWare. Probabilistic systems analysis and applied probability, 2010.
- University of Nairobi. Introduction to probability and statistics (sma 140), 2021. Accessed: 2024-09-09.
- Grimmett GR, Stirzaker DR. Probability and Random Processes. Oxford University Press, 2nd edition, 1992.